getwd()1 Learning Objectives
- Introduce
RandRStudio; - Work with scripts;
- Learn to import data.
2 Introduction
R is a powerful ‘object-oriented’ (i.e., data are assigned to named objects) programming language specifically designed for statistics. Although there are many software packages that can be used for statistical analysis (e.g., SPSS or GenStat), we have chosen R for C7041 Experimental Design and Analysis because it is designed to help people with no programming experience to perform sophisticated statistical analysis with minimum effort. Key features that make R so great include:
- Integrated data analysis and visualisation packages;
- Freely available and open-source;
- Widely used throughout industry and science;
- Continuously growing;
- Facilitates reproducible data analysis and research.
We also highly recommend download another piece of free software called RStudio. This software is an integrated development environment (IDE) for R that works with any standard version of R higher than 2.11. R itself does not have a graphical interface, but most people interact with R through graphical platforms that provide extra functionality. We will use RStudio as a graphical front-end to R, so that we can access our scripts and data, find help, and preview plots and outputs all in one place. Using RStudio will greatly enhance your programming experience and slightly lessen the learning curve.
3 Software Installation
To fully engage with the computer labs for C7041 Experimental Design and Analysis you will need to use R. You have three options for this:
- Download and install
Rfrom CRAN andRStudiofrom Posit. <- Preferred option! - Open a free
RStudio Cloudaccount and accessRthrough your web browser. - Open a free
Google Colaboratoryaccount and accessRthrough your web browser.
Regardless of your operating system, you should install R before installing RStudio. It is a good idea to go ahead and install the latest version of R if you have an older version installed.
4 RStudio
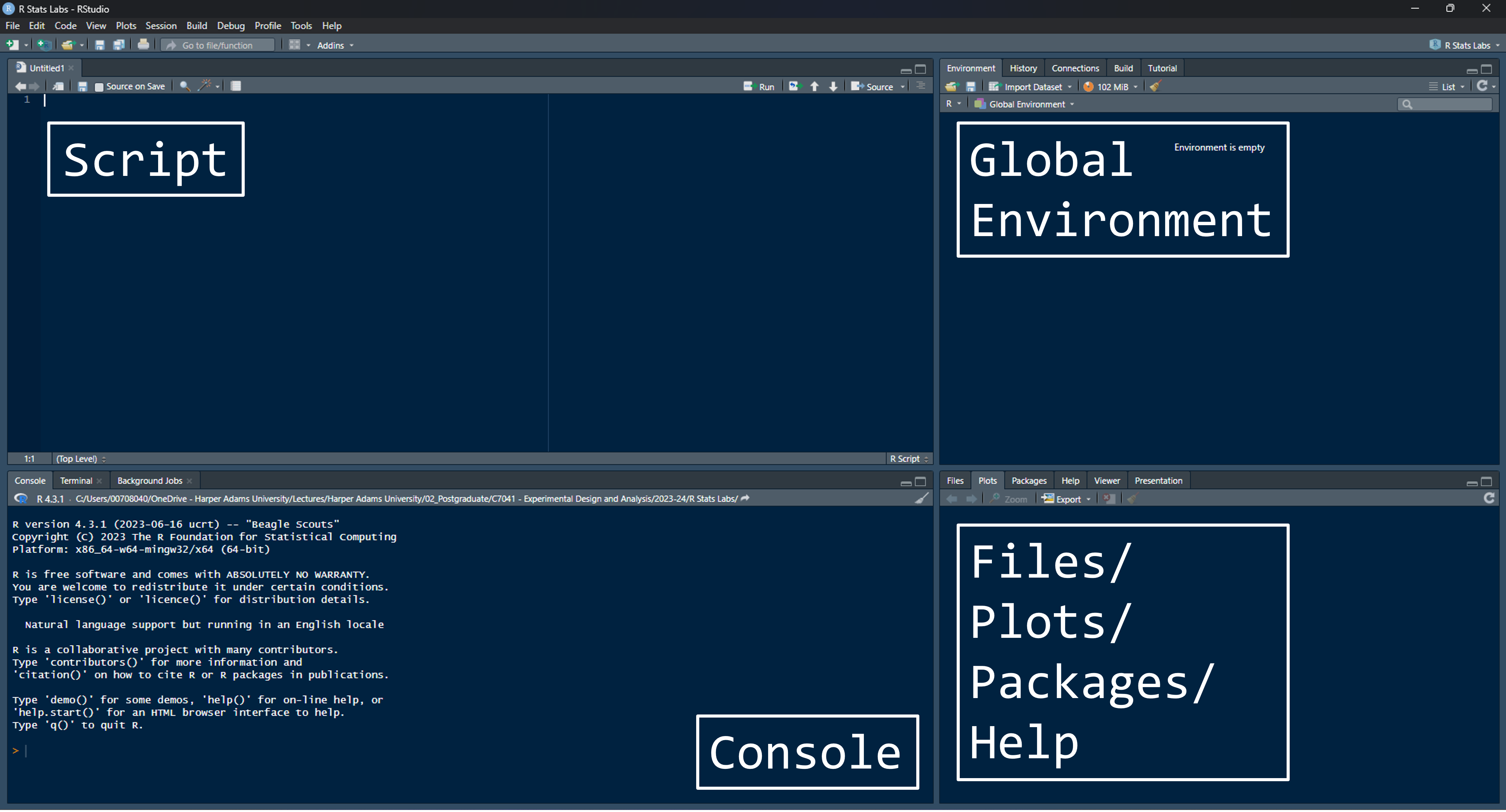
Once you have installed R and RStudio you can open RStudio and start using R. The first thing you will notice is that RStudio has a number of windows open by default. These windows are:
Script Window: The script window is located in the upper left of the RStudio console by default. You may need to open a script or start a new one: File > New File > R Script.
Console Window: The Console window is in the lower left by default. Notice there are several other tabs visible, but we will only mention the Console for now. The Console is the place where text outputs will be printed (e.g., the results of statistical tests), and also is a place where R will print Warning and Error messages.
Global Environment Window: The Global Environment is in the Environment tab in the upper right of RStudio by default. This pane is useful in displaying data objects that you have loaded and available.
Plots, etc. Window: The Plots window is a tab in the lower right by default. This is the place where graphics output is displayed and where plots can be named, resized, copied and saved. There are some other important tabs here as well, which you can also explore. When a new plot is produced, the Plots tab will become active. This also contains tabs to access other key information such as help.
5 R Scripts
5.1 What is a script?
A script is a plain text file that contains a series of R commands. Scripts are a great way to keep track of your work and to save your code for future use. You can also use scripts to create reproducible research. This means that you can share your code with others and they can reproduce your results exactly. This is a great way to make your research more transparent and reproducible.
5.2 Creating a script
To create a new script, go to File > New File > R Script. This will open a new window in the top left of RStudio where you can type your code. You can also open an existing script by going to File > Open File and selecting the script you want to open.
5.3 Script organisation
A goal is that your scripts should contain only important R commands and information, in an organized and logical way that has meaning for other people, maybe for people you have never spoken to. A typical way to achieve this is to organize every script according to the same plan. This is a plan that we recommend:
- Header
- Contents
- Code chunks
5.3.1 Header
The header is the first part of the script. It contains information about the script, such as the title, author, date, and a description of the script. The header is important because it provides information about the script and its purpose. The header is also a great place to include a description of the script. This is useful if you are sharing your script with others and want to explain what it does.
# A typical script Header
## HEADER ####
## Who: <your name>
## What: My first script
## Last edited: yyyy-mm-dd (ISO 8601 date format... Google it!)
####5.3.2 Contents
You may want to include a contents section near the top to provide a ‘road map’ for your script & analysis. For example:
# A typical script Contents section
## CONTENTS ####
## 00 Setup
## 01 Graphs
## 02 Analysis
## 03 Etc5.3.3 Code chunks
A ‘Code chunk’ break is just a notation method used to aid the readability of the script and to provide a section for each item in your table of contents. A code chunk is just a section of code set off from other sections.
Below is the beginning of a typical code chunk in an R script.
- Code chunks must start with at least one hash sign
#; - Should have a title descriptive of code chunk contents;
- End with (at least) four hash signs
####; - Consecutively numbered titles can make things very tidy.
For example:
## 01 This here is the first line of MY CODE CHUNK ####5.3.5 Running code
To run code in a script, you can either highlight the code you want to run and click the Run button in the top right of the script window, or you can use the keyboard shortcut Ctrl + Enter (Windows) or Cmd + Enter (Mac). You can also run the entire script by clicking the Source button in the top right of the script window, or by using the keyboard shortcut Ctrl + Shift + S (Windows) or Cmd + Shift + S (Mac).
6 Working Directories
R does not have omniscient knowledge of your computer and its contents. It will look for files and write new files by default to wherever its current working directory is set. If you open a blank R studio (or blank R GUI) instance it will default to wherever is set in the global options: typically your local home directory. Changing the location of the files pane in Rstudio does not change the working directory and so you may have difficulty loading data files, or finding any figures or outputs you have saved. Another great feature of the Rproject approach which means you can share projects between computers, and even between operating systems.
Generally speaking, if you use the in-built project manager within RStudio, then this issue of setting working directories should never arise for you. Instead, you can open a specific Rproject by double clicking on the *.Rproject file in a finder or folder browser window, or by choosing File, Open Project from the RStudio drop down menu, or via the icon in the top right corner of your RStudio application. To set up a new Rproject you should click on File > New Project > New Directory > New Project > Enter a Directory Name > Choose Directory Location.
To see your current directory execute the following code:
You can change working directory in any of these three ways:
Use the
setwd()function in Windows:setwd("C:/Users/User Name/Documents/FOLDER")or on macOS:
setwd("/Users/User Name/Documents/FOLDER")Use
Session>Set Working DirectoryUsing the
Filespane, navigate to the folder you want and then select the drop down menuMoreand thenSet as Working Directory.
7 R Packages
A package is a bundle of commands that can be loaded into R to provide extra functionality. For example, you might load a package for formatting data, or for making maps. There are thousands of packages available for R, and you can even write your own packages. Packages are a great way to extend the functionality of R and to make your code more efficient. To install a package, type install.packages("package-name"). You only need to install packages once, so in this case you can type directly in the console box, rather than saving the line in your script and re-installing the package every time.
Once installed, you just need to load the packages using library(package-name). You will need to load the package every time you start a new R session, so it is a good idea to include this line in your script. You can load multiple packages at once by separating them with commas (e.g., library(package1, package2, package3)).
# Install packages
install.packages("tidyverse")
# Load packages
library(tidyverse)8 Import and Check Data
Practice is the best way to learn any new language, so let’s jump straight in. We will start by importing some data into R. There are a very large number of ways to do this and most people eventually find their own workflow. We think it is best for most people to use Excel (.xlsx) or comma separated values (.CSV) files in Tidy Data format.
The basics of reading external files from a script is to to use the read.xlsx() function in the openxlsx package (you will probably need to install this with the install.packages() function), or else to use read.csv() that comes standard in base R. We will briefly try both.
8.1 Import file using read.xlsx()
# Get working directory
getwd() # Prints working directory in Console
# Set working directory
setwd("C:/Users/User Name/Documents/FOLDER") # Change to your directory
# Install package
install.packages("openxlsx", dep = TRUE) # Install package
# Load package
library(openxlsx) # Load package
# Import data
data <- read.xlsx("intro_data_1.xlsx", # File name
sheet = 1, # Sheet number
colNames = TRUE, # Column names
detectDates = TRUE) # Detect dates8.2 Import file using read.csv()
The same procedure works with comma separated values data files, and other kinds of files that you want to read into R, except that the R function used will be specific to the file type. For example, read.csv() for CSV files, read.delim() for TAB delimited files, or read.table() as a generic function to tailor to many types of plain text data files (there are many others, but this is enough for now).
# Get working directory
getwd() # Prints working directory in Console
# Set working directory
setwd("C:/Users/User Name/Documents/FOLDER") # Change to your directory
# Import data
data <- read.csv("intro_data_1.csv", # File name
header = TRUE) # Column names8.3 Check data
A really important step is to check that your data was imported without any mistakes. It’s good practice to always run this code and check the output in the console - do you see any missing values, do the numbers/names make sense? If you go straight into analysis, you risk later finding out that R didn’t read your data correctly and having to re-do it, or worse, analysing wrong data without noticing. To preview more than just the few first lines, you can also click on the object in your Environment panel, and it will show up as a spreadsheet in a new tab next to your open script. Large files may not display entirely, so keep in mind you could be missing rows or columns.
head(data) # Displays the first few rows id aphid treatment rt peak.area conc.tot conc.ind
1 CG19-14 Brevicoryne Plant 16.247 154123 9.375757 3.125252
2 CG19-23 Brevicoryne Plant 16.240 77839 4.735176 1.578392
3 CG19-24 Brevicoryne Plant 16.243 204238 12.424400 4.141467
4 CG19-09 Myzus Plant 16.241 4783237 290.978422 96.992807
5 CG19-13 Myzus Plant 16.245 2653330 161.409894 53.803298
6 CG19-26 Myzus Plant 16.240 3224152 196.134680 65.378227tail(data) # Displays the last rows id aphid treatment rt peak.area conc.tot conc.ind
13 CG19-15 Myzus AD+SA 16.243 4034920 245.456090 81.818697
14 CG19-18 Myzus AD+SA 16.242 2442438 148.580711 49.526904
15 CG19-22 Myzus AD+SA 16.242 5650064 343.710066 114.570022
16 CG19-17 Myzus AD-SA 16.242 51450 3.129855 1.043285
17 CG19-20 Myzus AD-SA 16.242 846354 51.486211 17.162070
18 CG19-21 Myzus AD-SA 16.244 110176 6.702331 2.234110str(data) # Tells you whether the variables are continuous, integers, categorical or characters'data.frame': 18 obs. of 7 variables:
$ id : chr "CG19-14" "CG19-23" "CG19-24" "CG19-09" ...
$ aphid : chr "Brevicoryne" "Brevicoryne" "Brevicoryne" "Myzus" ...
$ treatment: chr "Plant" "Plant" "Plant" "Plant" ...
$ rt : num 16.2 16.2 16.2 16.2 16.2 ...
$ peak.area: num 154123 77839 204238 4783237 2653330 ...
$ conc.tot : num 9.38 4.74 12.42 290.98 161.41 ...
$ conc.ind : num 3.13 1.58 4.14 96.99 53.8 ...str(object.name) is a great command that shows the structure of your data. So often, analyses in R go wrong because R decides that a variable is a certain type of data that it is not. For instance, you might have four study groups that you simply called “1, 2, 3, 4”, and while you know that it should be a categorical grouping variable (i.e., a factor), R might decide that this column contains numeric (numbers) or integer (whole number) data. If your study groups were called “one, two, three, four”, R might decide it’s a character variable (words or strings of words), which will not get you far if you want to compare means among groups. Bottom line: always check your data structure!
You will notice that both the aphid and treatment variables are characters. This is because R has no way of knowing that these are categorical variables. We will need to convert them to factors before we can do any analysis.
# Convert variables to factors
data$aphid <- as.factor(data$aphid)
data$treatment <- as.factor(data$treatment)In that last line of code, the as.factor() function turns whatever values you put inside into a factor (here, we specified we wanted to transform the character values in the aphid and treatment columns from the data object). However, if you were to run just the bit of code on the right side of the arrow, it would work that one time, but would not modify the data stored in the object. By assigning with the arrow the output of the function to the variable, the original data$aphid and data$treatment variables are overwritten (i.e.,) the transformation is stored in the object.
9 Accessing Help
R has a very extensive help system. You can access the help system by typing ? followed by the name of the function you want help with. For example, to get help with the read.csv() function, type ?read.csv. You can also use the help() function, for example help(read.csv). You can also use the help.search() function to search for help on a particular topic, for example help.search("import data").
10 Activities
10.1 Import data
Import the data file intro_data_1.xlsx into R using the read.xlsx() function in the openxlsx package. The data file is located in the data folder in the R project directory. You will need to first download some data from here.
💡 Click here to view a solution
# Install package
#install.packages("openxlsx", dep = TRUE)
# Load package
library(openxlsx)
# Import data
my_data <- read.csv("data/intro_data_2.csv",
header = TRUE)10.2 Check data
Check the data using the head(), tail() and str() functions. What do you notice about the taxonGroup variable? What do you think the problem is? Fix the problem!
💡 Click here to view a solution
# View data
str(my_data) # taxonGroup is a character variable'data.frame': 25684 obs. of 5 variables:
$ organisationName: chr "Joint Nature Conservation Committee" "Joint Nature Conservation Committee" "Joint Nature Conservation Committee" "British Trust for Ornithology" ...
$ gridReference : chr "NT265775" "NT235775" "NT235775" "NT27" ...
$ year : int 2000 2000 2000 2000 2000 2001 2001 2001 2001 2001 ...
$ taxonName : chr "Sterna hirundo" "Sterna hirundo" "Sterna paradisaea" "Branta canadensis" ...
$ taxonGroup : chr "Bird" "Bird" "Bird" "Bird" ...# Convert to factor
my_data$taxonGroup <- as.factor(my_data$taxonGroup)
# View data
class(my_data$taxonGroup) # taxonGroup is now a factor variable[1] "factor"Why are character variables a problem?
💡 Click here to view an answer
Because R will treat them as continuous variables, and you will not be able to do any meaningful analysis with them.
5.3.4 Comments
Comments are lines of code that are not executed by
R. They are used to provide information about the code. Comments are useful for explaining what the code does and why it is there. Comments are also useful for providing information about the code that is not obvious from reading the code itself. For example, you may want to explain why you chose a particular statistical test or why you chose a particular variable to include in your analysis. Comment lines begin with the # character and are not treated as “code” byR: